-
- Downloads
Updating Examples
Showing
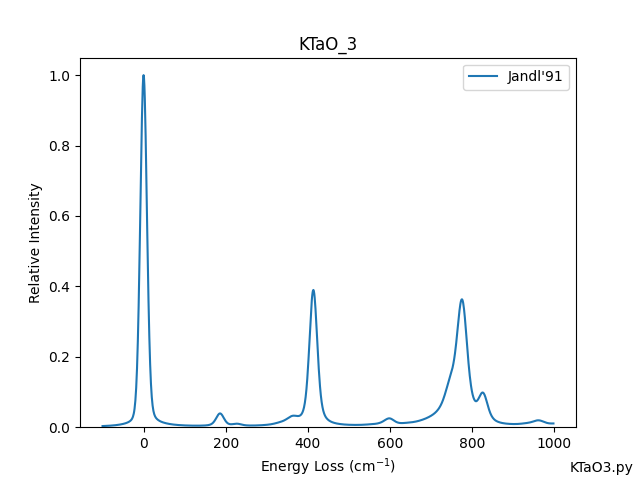
- Examples/KTaO3.png 0 additions, 0 deletionsExamples/KTaO3.png
- Examples/KTaO3.py 6 additions, 0 deletionsExamples/KTaO3.py
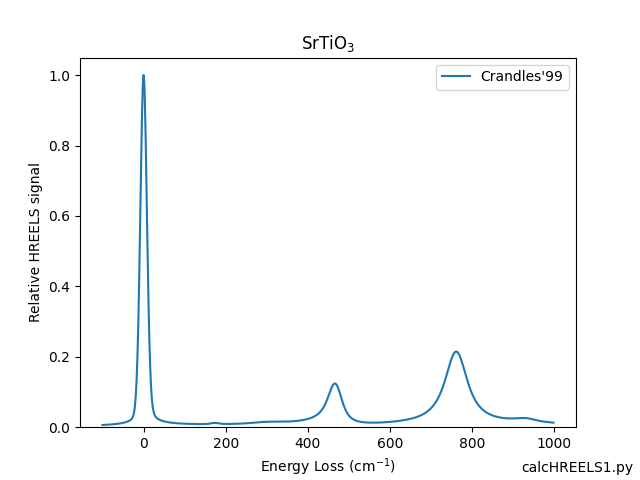
- Examples/calcHREELS1.png 0 additions, 0 deletionsExamples/calcHREELS1.png
- Examples/calcHREELS1.py 6 additions, 0 deletionsExamples/calcHREELS1.py
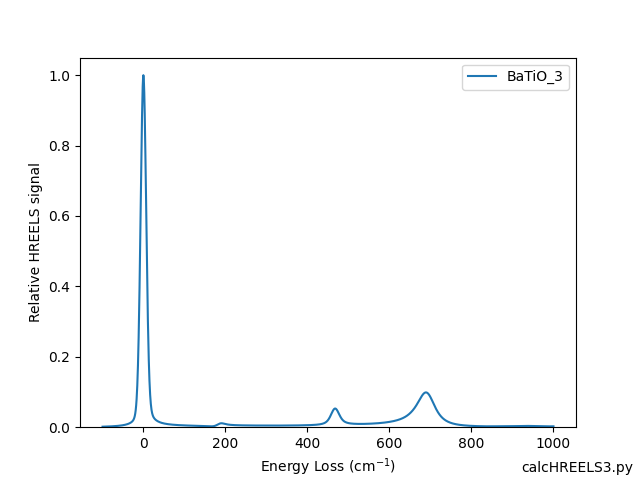
- Examples/calcHREELS3.png 0 additions, 0 deletionsExamples/calcHREELS3.png
- Examples/calcHREELS3.py 8 additions, 2 deletionsExamples/calcHREELS3.py
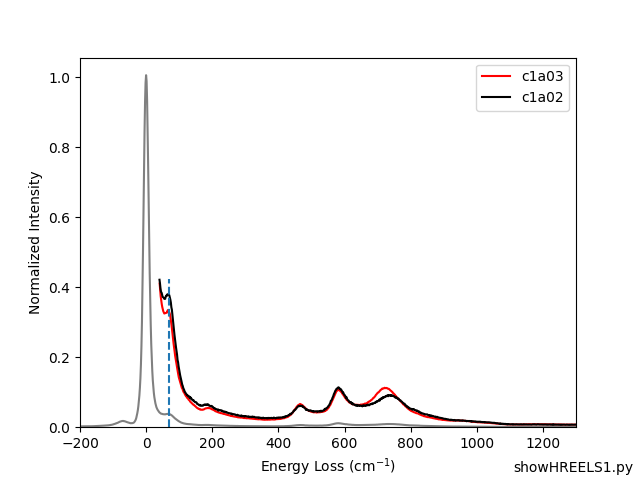
- Examples/showHREELS1.png 0 additions, 0 deletionsExamples/showHREELS1.png
- Examples/showHREELS1.py 7 additions, 2 deletionsExamples/showHREELS1.py
- Examples/showHREELS2.png 0 additions, 0 deletionsExamples/showHREELS2.png
- Examples/showHREELS2.py 7 additions, 0 deletionsExamples/showHREELS2.py
- libhreels/HREELS.py 4 additions, 2 deletionslibhreels/HREELS.py
- libhreels/calcHREELS.py 6 additions, 5 deletionslibhreels/calcHREELS.py
Examples/KTaO3.png
0 → 100644
29.4 KiB
Examples/calcHREELS1.png
0 → 100644
28.3 KiB
Examples/calcHREELS3.png
0 → 100644
24.6 KiB
Examples/showHREELS1.png
0 → 100644
30.7 KiB
Examples/showHREELS2.png
0 → 100644
37.1 KiB